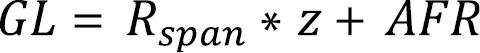(C) 2013 Michela Pacifici. This is an open access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC-BY), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
Citation: Pacifici M, Santini L, Di Marco M, Baisero D, Francucci L, Grottolo Marasini G, Visconti P, Rondinini C (2013) Generation length for mammals. Nature Conservation 5: 87–94. doi: 10.3897/natureconservation.5.5734 Resource ID: Dryad key: 10.5061/dryad.gd0m3
Resource citation: Pacifici M, Santini L, Di Marco M, Baisero D, Francucci L, Grottolo Marasini G, Visconti P, Rondinini C (2013) Database on generation length of mammals. 5427 data records. Online at http://doi.org/10.5061/dryad.gd0m3, version 1.0 (last updated on 2013-08-27), Resource ID: 10.5061/dryad.gd0m3, Data Paper ID: doi: 10.3897/natureconservation.5.5734
Generation length (GL) is defined as the average age of parents of the current cohort, reflecting the turnover rate of breeding individuals in a population. GL is a fundamental piece of information for population ecology as well as for measuring species threat status (e.g. in the IUCN Red List). Here we present a dataset including GL records for all extant mammal species (n=5427). We first reviewed all data on GL published in the IUCN Red List database. We then calculated a value for species with available reproductive parameters (reproductive life span and age at first reproduction). We assigned to missing-data species a mean GL value from congeneric or confamilial species (depending on data availability). Finally, for a few remaining species, we assigned mean GL values from species with similar body mass and belonging to the same order. Our work provides the first attempt to complete a database of GL for mammals; it will be an essential reference point for all conservation-related studies that need pragmatic information on species GL, such as population dynamics and applications of the IUCN Red List assessment.
Age at first reproduction, conservation assessment, IUCN Red List, longevity, reproductive life span
Generation length (GL) has been defined in a number of ways and has been approximated with a number of different formulas (
This database covers all 5427 extant species in the class Mammalia. The taxonomy follows the IUCN Red List of Threatened Species version 2012.2.
For 439 species, we used stated GL in years available from published IUCN Red List assessments (
For the mammal body masses, we used PanTHERIA (
For 315 species, lacking a congeneric or confamilial species in the same bin of log body mass, we assigned the mean GL value of congenerics or confamilials, irrespective of their body mass. For the remaining species (n=116, 2.1 % of the total), where no information was available for congeneric or confamilial species, we assigned the mean GL value of species in the same bin of log body mass, belonging to the same order, or simply the mean GL values of the order (2 species, Ptilocercus lowii and Cyclopes didactylus). We made an exception for the two species of Dermoptera and 9 species of small mammals (body mass < 100 g); since they were the only representatives of their orders, we estimated mean GLs from species belonging to the same bin of log body mass. In this way, we obtained a GL value for all existing 5427 mammals.
We estimated GL for mammals from information on species age at first reproduction and reproductive life span, by applying the methodology described in the IUCN Red List Guidelines (
where Rspan is the species reproductive life span, calculated as the difference between the age at last reproduction and the age at first reproduction (AFR), and z is a constant “depending on survivorship and relative fecundity of young vs. old individuals in the population” (
Since age at last reproduction is generally related to longevity in the wild (
For carnivore and ungulate species, we completed missing data on maximum longevity and age at sexual maturity through a multiple imputation procedure (
The dataset includes generation lengths for 5427 mammal species. Fields given are:
TaxID: identification number of species;
Order;
Family;
Genus;
ScientificName;
AdultBodyMass_g: body mass of species in grams;
Sources_AdultBodyMass: AnAge, Animal Diversity, Encyclopedia of Life (eol.org/),
Nowak and Paradiso 1999 , PanTHERIA,Smith et al. 2003 ,Verde Arregoitia et al. 2013 , Mean congenerics, Mean_confamilials;Max_longevity_d: maximum longevity (days) mediated from PanTHERIA, AnAge and Carn_Ung (multiple imputation for carnivores and ungulates);
Sources_Max_longevity: AnAge, Carn_ung (multiple imputation for ungulates and carnivores) and PanTHERIA;
CalculatedRspan_d: reproductive life span (days) calculated from maximum longevity and age at first reproduction;
AFR_d; age at first reproduction (days);
Data_AFR: calculated or published data;
CalculatedGL_d: GL (days) calculated from reproductive life span and age at first reproduction;
GenerationLength_d: best known estimate of GL (days), including information taken from IUCN database, calculated data and mean estimates;
Sources_GL:
GMA (IUCN Red List data);
Rspan-AFB (GL calculated as the difference between reproductive life span and age at first birth);
Rspan-AFR(SM+Gest) (when data on age at first reproduction were not available, we calculated this parameter as the sum between age at female sexual maturity and gestation length);
Rspan-ASMmales (GL calculated with age at sexual maturity for males, when data on age at first reproduction for females were not available);
Mean_congenerics_same_body_mass (mean GL calculated from congeneric species in the same bin of log body mass);
Mean_congenerics (mean GL calculated from congeneric species, irrespective of body mass);
Mean_family_same_body_mass (mean GL calculated from confamilial species in the same bin of log body mass);
Mean_family (mean GL calculated from confamilial species, irrespective of body mass);
Mean_order_same_mass (for species with unknown parameter estimates, we assigned the mean GL value of species in the same bin of log body mass and belonging to the same order);
Mean_order (mean GL calculated from species belonging to the same order, irrespective of body mass);
Mean_all_orders_same_body_mass (species for which we estimated mean GL from species belonging to the same bin of log body mass).
The data underpinning the analysis reported in this paper are deposited in the Dryad Data Repository at http://doi.org/10.5061/dryad.gd0m3
Pacifici M, Santini L, Di Marco M, Baisero D, Francucci L, Grottolo Marasini G, Visconti P, Rondinini C (2013) Generation length for mammals. Nature Conservation 5: 87–94. doi: 10.3897/natureconservation.5.5734
Database on generation length of mammals. (doi: 10.3897/natureconservation.5.5734.app). File format: Microsoft Excel file (xls).
Explanation note: Database on generation length of all extant mammals, with 5427 records.